Abstract
Single cell analysis tools are crucial to understand the role that rare or heterogeneous cancer cells play in tumor progression. To enable the characterization of genetic variation within cancer cell populations, we developed a novel approach that barcodes amplified genomic DNA of individual cells confined to microfluidic droplets. The barcodes are used to reassemble the genetic profiles of individual cells from next-generation sequencing data. A key feature of our approach is the "two-step" microfluidic workflow. The microfluidic workflow first encapsulates individual cells in droplets, lyses the cells and prepares the genomic DNA for amplification with proteases. Following this lysate preparation step, the proteases are inactivated and droplets containing the genomes of individual cells are then paired with molecular barcodes and PCR reagents. We demonstrate that the two-step microfluidic approach is superior to workflows without the two-step process for efficient DNA amplification on thousands of individual cells per run with high coverage uniformity and low allelic dropout of targeted genomic loci.
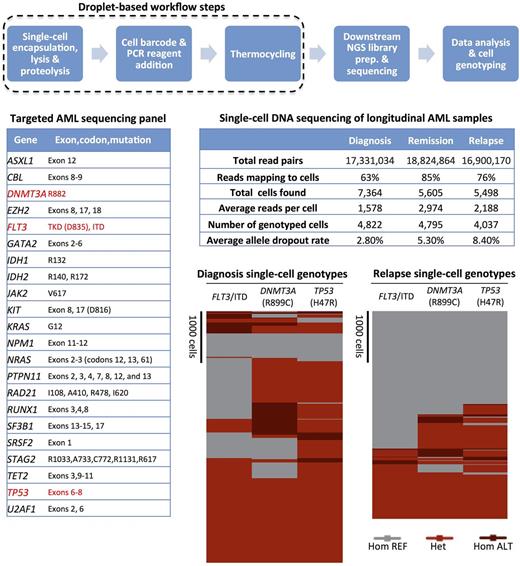
To apply our single-cell sequencing technology to human tumor samples, we developed a targeted panel to partially sequence 23 genes frequently mutated in acute myeloid leukemia (AML) including TP53 , DNMT3A , FLT3 , NPM1 , NRAS , IDH1 and IDH2 . Using this panel, we were able to sensitively assay SNP and indel defined clones within AML samples collected longitudinally at the time of diagnosis, remission and relapse. Our single-cell data indicates that clonal populations inferred from VAFs obtained from bulk sequencing data may not fully resolve the heterogeneity within tumors; moreover, the single-cell nature of our approach enabled the unambiguous colocalization of multiple mutations within subclones not possible with bulk measurements. Collectively, our results show a greater degree of heterogeneity in AML tumor samples than is commonly appreciated with traditional sequencing paradigms and they demonstrate the value of single-cell analysis for AML.
Eastburn: Mission Bio: Employment, Equity Ownership. Pellegrino: Mission Bio: Employment, Equity Ownership. Sciambi: Mission Bio: Employment, Equity Ownership. Treusch: Mission Bio: Employment, Equity Ownership. Gokhale: Mission Bio: Employment, Equity Ownership. Jacob: Mission Bio: Employment, Equity Ownership. Chen: Mission Bio: Employment, Equity Ownership. Jones: Mission Bio: Employment, Equity Ownership. Takahashi: Symbio Pharmaceuticals: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal